Voxelcore Readme
Pipeline
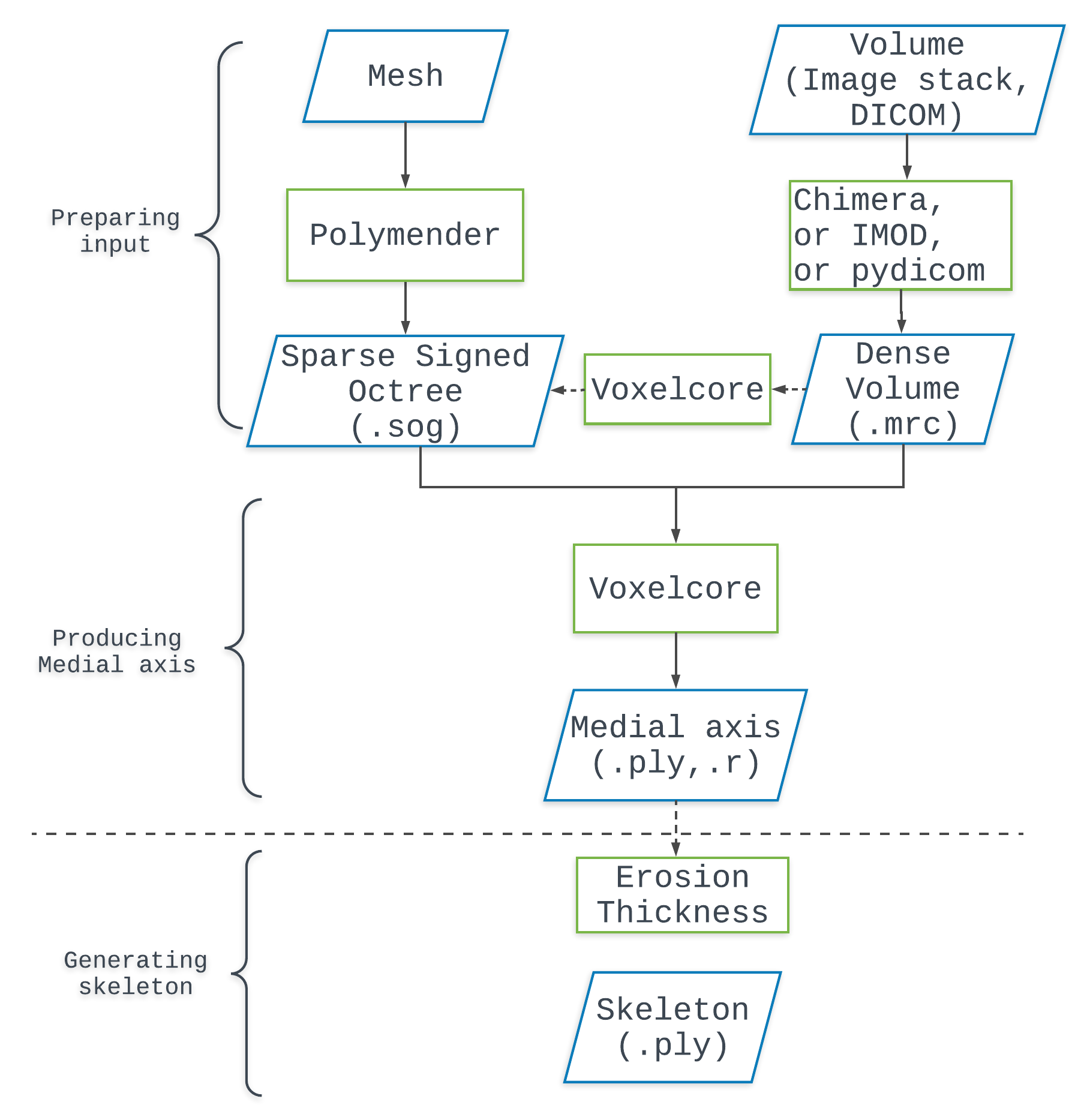
Usually the user is in pocession of a digital shape that’s either represented as a volume by a set of images, or as a surface by a triangular mesh. Below we explain how to convert these datasets into a dense volume (.mrc) or a sparse signed octree (.sog) respectively, both are acceptable input formats to our program
Preparing input
As shown in Figure 1, input to our program shall be prepared differently depending on the type of original dataset.
From image stack to .mrc
If the user has a stack of images (e.g. png, tiff), there exists three approaches to convert that into a volume in format .mrc, which can be input to our voxelcore program. A .mrc file stores a 3D volume where each voxel has a density value. Voxelcore assumes that object voxels have values > 0, while background voxels <= 0.
- Interactive programs Free-wares such as Paraview and Chimera can open stack of images in common formats (e.g. tiff, png, jpg), visualizing a volume dataset within the program. This voluem can be then saved as a
.mrcfile. This approach is very easy to perform, but may not be suited for purpose of batch processing. - Command line utilities One can easily find a handful command line tools online that process images into volume file. However to our knowledge, these utilities only accept certain image formats. For example the IMOD package offers a command line program called
tif2mrcthat converts a stack of tiff images to a mrc file. The user would need to convert their images to tiff first to use this utility. This approach is easy to batch process any number of datasets, but no one utility can cover a wide range of formats. - Scripts The 3rd option is to take a bit self initiative to write some scripts to do the job. Though myriad scripting languages are out there, we had the best experience with python, as the vastly available modules help pull the work off more easily. For example, pydicom can easily parse DICOM files, a powerful yet sophisticated medical image format. Then the content can be written to a
.mrcfile using another python library, mrcfile. Similar library exists that parse other widly used image formats. These specialized python scripts can be further wrapped by a format dispatcher that can better automate the task. The advantage is apparently an automatic conversion tool suitable for batch processing a variety of formats the user wants to support.
From mesh to .sog
On the other hand, users with a mesh representing the shape are suggested to convert it to a sparse signed octree in format .sog.
- Polymender can be used to produce a SOG file from the given triangular mesh (route 2 in Figure 1) (see Polymender, or download the program and see the readme.txt).
- Voxelcore program itself can produce a SOG file from a MRC file in mode
mrc2sog:voxelcore -md=mrc2sog -mrc=mrc_file_path -sog=sog_output_pathThe original mrc can be deleted if the user has no other intention with it, since the sog file contains complete information about the volume.
Producing medial axis
Now the input is ready, our program can be called in the following format
voxelcore -md=validmode
The program runs in a few modes to help generate the medial axis of the input shape, and other relevant information.
Generating medial axis from volume with -md=vol2ma. In this mode, the program will read in the specified volume file, and output the voxel core to approximate true medial axis. Complete information about this mode is listed below (can be printed by typing voxelcore -md=vol2ma).
voxelcore -md=vol2ma <in: volume fullpath. ext: .mrc/.sog> <out: MA fullpath. ext: .ply> [optional args]
optional:
-tt (1 or more comma-sep lambda pruning thresholds. OPTIONAL.) type: string default: "0.04"
-fullOrPruned (write full or prunned voxel core (0: pruned, 1: full, 2: both). OPTIONAL.) type: int32 default: 0
Generating voxel shape boundary with -md=vol2mesh. In this mode, the boundary of the voxel shape represented by a volume file is extracted as a .off mesh file. Special argument can be specified to only output a point cloud (without faces). Full information about this mode is as follows (can be printed by typing voxelcore -md=vol2mesh).
voxelcore -md=vol2mesh <input vol fullpath> <output mesh fullpath(w/o extension)> [optional args]
optional:
-onlyBndryVts (only extract the vertices of the voxel boundary. OPTIONAL.) type: bool default: true
-write2node (write voxel boundary vts to .node file in input file folder. OPTIONAL.) type: bool default: false
Converting .mrc to .sog with -md=mrc2sog. A .mrc volume file can be converted to a .sog volume file in this mode (as depicted in the pipeline figure by a dotted arrow). Again typing voxelcore -md=mrc2sog prompts the full information abou this mode.
voxelcore -md=vol2mesh <input vol fullpath> <output mesh fullpath(w/o extension)> [optional]
optional:
-onlyBndryVts (only extract the vertices of the voxel boundary. OPTIONAL.) type: bool default: true
-write2node (write voxel boundary vts to .node file in input file folder. OPTIONAL.) type: bool default: false